(A01-2)
Nuclear Magnetic Resonance (NMR) spectroscopy is one of the most powerful and robust measurement methods that are able to non-invasively observe the structural dynamics of molecules. Recent advances in NMR hardware technology enable us to measure complex biomolecules such as proteins, however, the size of measured data is huge, measurement time is lengthy, obtained spectrum is complex, and its analysis is complicated. In addition, structure calculation of biomolecule is time-consuming complicated process because the large amounts of data are required. In this project, focusing on the sparseness of NMR data and structure information of biomolecule, we aim to achieve faster and more precise NMR data measurement and analysis of biomolecule by solving the issues arising from complexity and complicatedness of NMR analysis with the sparse modeling, as shown in Figure 1.
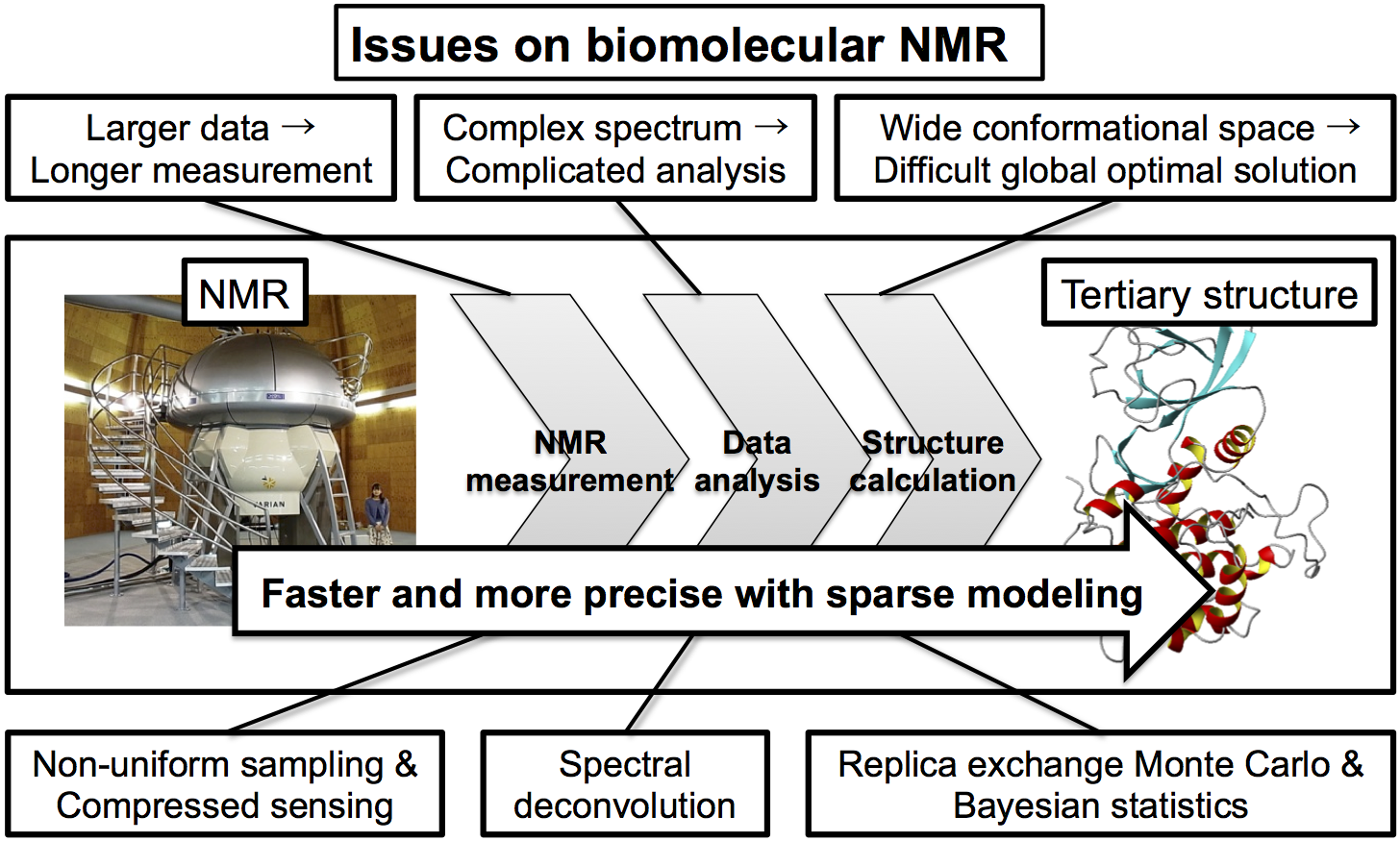
Figure 1
In NMR measurements on complex biomolecules such as proteins and nucleic acids, most of them are difficult to be analyzed because it is unstable enough to be damaged during long measurement time resulted by required data amount. Therefore, the development of a method for obtaining high resolution NMR data in a short period of time is desired. Recently, the method to shorten the measurement time of NMR signal, which is generally recorded with uniform sampling, has been proposed by recording reduced number of data sets with non-uniform sampling and interpolating them with maximum entropy or compressed sensing methods. In addition, the data analysis with high precision is difficult to be achieved due to the difficulty of peak identification and discrimination on the complicated NMR spectrum, and, on the structure calculation based on the analyzed data, the difficulty in obtaining optimal solution has been problematic because it is usually trapped in local minima due to the difficulty to search over the broad ranges of conformational space. Our experience and results obtained from the project on the large-scale structure analysis of biomolecules, a.k.a. "Structural genomics", using NMR spectroscopy and the development of de facto standard software for structure calculation based on NMR data led us to the idea that, with sparse modeling, faster and more precise NMR data measurement and analysis can promote progress in NMR measurement on complex biomolecular systems.
Focusing on the sparseness of NMR data and structure information of biomolecules, this planned research project aims to promote progress in NMR measurement on complex biomolecular systems by achieving faster and more precise NMR data measurement and analysis of biomolecule with sparse modeling, especially addressing the following three research subjects.
- [Subject 1] Faster and more precise NMR measurement
Dramatic shortening of measurement time and improvement of the spectrum quality will be achieved by obtaining NMR signal in a sparsely sampled manner and interpolating them with sparse modeling. - [Subject 2] Faster and more precise NMR data analysis
NMR data analysis will become faster and more precise by improving peak discrimination and identification on complicated NMR spectra and further automatizing the processes. - [Subject 3] Faster and more precise NMR structure calculation
We will establish an algorithm for determining parameters in rational and quantitative way by data analysis based on Bayesian model in order to obtain global optimum solution with replica exchange Monte Carlo method, enabling faster and more precise NMR structure calculation.
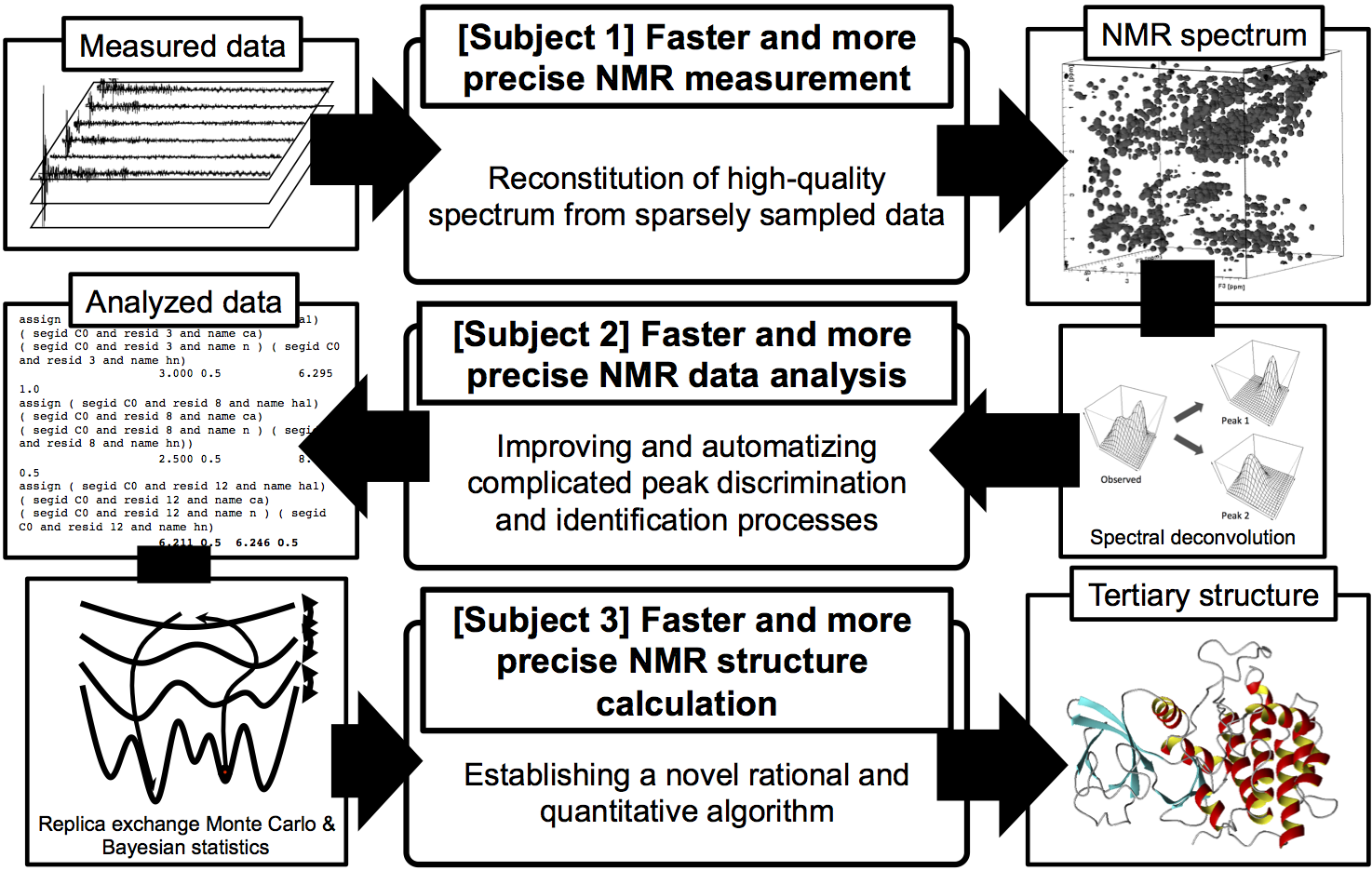
Figure 2